In a recent study posted to the bioRxiv* pre-print server, researchers at Cornell University College of Veterinary Medicine and the University of Illinois Urbana-Champaign compared the infection dynamics, pathogenicity, and tissue tropism of multiple severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants in adult cats.
A new wave of SARS-CoV-2 breakthrough infections is occurring around the world due to emerging new SARS-CoV-2 variants of concern (VOCs). Hence, a better understanding of their infectivity and pathogenesis is critical for the development of improved vaccines and therapeutics against novel SARS-CoV-2 VOCs.
 Study: The Omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and Delta variants in a feline model of SARS-CoV-2 infection. Image Credit: NIAID
Study: The Omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and Delta variants in a feline model of SARS-CoV-2 infection. Image Credit: NIAID
About the study
In the present study, researchers conducted a 14-day experiment with SARS-CoV-2-infected adult cats in the animal biosafety level 3 (ABSL-3) facility at Cornell University in New York, United States. They infected highly susceptible cats with SARS-CoV-2 D614G (B.1) strain, and two VOCs, viz., Delta (B.1.617.2) and Omicron BA.1.1 (B.1.1.529).
The researchers first assessed the infected cats for clinical parameters, including body weight and clinical signs of respiratory disease. They collected their nasal and oropharyngeal secretions (OPS) and rectal swabs (RS) to detect the presence of SARS-CoV-2 ribonucleic acid (RNA) and infectious viral particles via real-time reverse transcriptase-polymerase chain reaction (RT-PCR). Likewise, the team assessed the viral RNA load and infectious virus titers in bronchoalveolar lavage fluid (BALF) of the test animals.
The researchers extracted several organs, including nasal turbinate, tonsil, lungs, trachea, liver, spleen, heart, kidney, small intestine, and associated lymph nodes from infected test animals to locate the sites of SARS-CoV-2 replication and assess tissue tropism. Next, they performed quantitative RT-PCR targeting the envelope (E) gene to quantify subgenomic RNA (sg-RNA) in the tissues of infected animals. Further, they used in situ hybridization and immunofluorescence (IFA) staining to assess the tissue distribution of SARS-CoV-2 in the respiratory tract and associated lymphoid tissues. The team also performed the histological examination of feline tissues collected on days three, five, and 14 from D614G-, Delta-, and Omicron-infected cats.
Finally, the team assessed the antibody responses to SARS-CoV-2 infection in cats using an indirect enzyme-linked immunosorbent assay (ELISA). In addition, they used a multiplex Luminex bead-based immunoassay to measure serum cytokine and chemokine levels throughout the SARS-CoV-2 infection window.
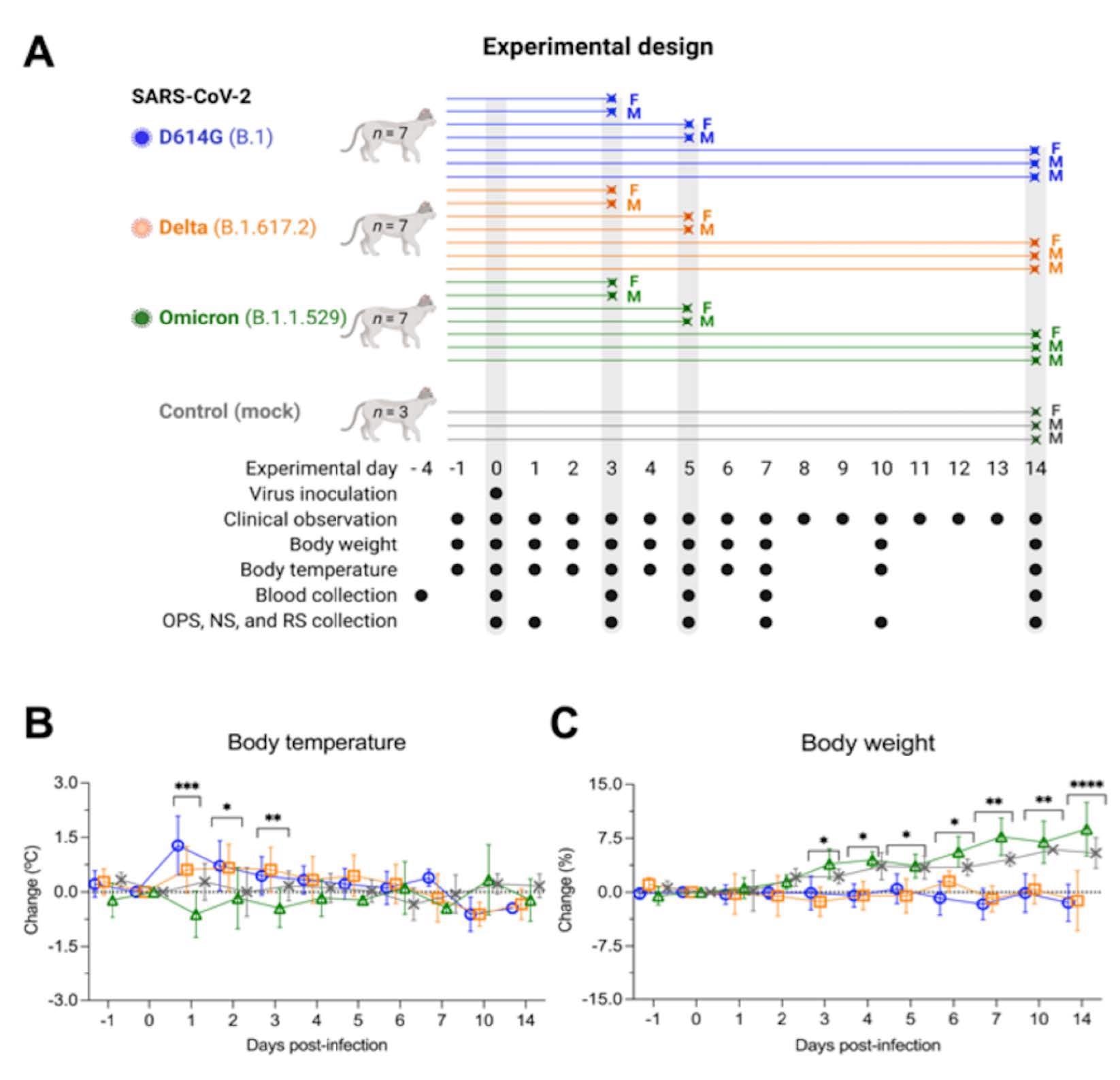 Experimental design, body temperature and weight changes following SARS-CoV-2 inoculation in cats. Schematic representation of the experimental design of infection study in adult cats (24-40-month-old) males and females. Animals were allocated in three inoculated groups (n = 7 per group) and a control group (mock-inoculated) (n = 3). Animals were inoculated intranasally with 1 ml (0.5 ml per nostril) of virus suspension containing 5 x 105 PFU of SARS-CoV-2 D614G (B.1), Delta (B.1.617.2), or Omicron BA.1.1 (B.1.1.529), or 1 ml of cell culture supernatant media (control mock-inoculated). On day 3 and 5 post-infection (pi), two cats from each group (one female and one male) were humanely euthanized and the remaining cats (one female and two males) were maintained until day 14 pi. Respiratory secretion, feces and serum were collected on the specific days as indicated (A). NS = nasal swab; OPS = oropharyngeal swab; RS = rectal swab; F = female; M = male. Body temperature (B) and body weight changes (C) following intranasal SARS-CoV-2 D614G (B.1), Delta (B.1.617.2), or Omicron BA.1.1 (B.1.1.529) inoculation throughout the 14-day experimental period. Data are presented as means ± standard deviation. * p < 0.05; ** p < 0.01; *** p < 0.005; **** p < 0.0001.
Experimental design, body temperature and weight changes following SARS-CoV-2 inoculation in cats. Schematic representation of the experimental design of infection study in adult cats (24-40-month-old) males and females. Animals were allocated in three inoculated groups (n = 7 per group) and a control group (mock-inoculated) (n = 3). Animals were inoculated intranasally with 1 ml (0.5 ml per nostril) of virus suspension containing 5 x 105 PFU of SARS-CoV-2 D614G (B.1), Delta (B.1.617.2), or Omicron BA.1.1 (B.1.1.529), or 1 ml of cell culture supernatant media (control mock-inoculated). On day 3 and 5 post-infection (pi), two cats from each group (one female and one male) were humanely euthanized and the remaining cats (one female and two males) were maintained until day 14 pi. Respiratory secretion, feces and serum were collected on the specific days as indicated (A). NS = nasal swab; OPS = oropharyngeal swab; RS = rectal swab; F = female; M = male. Body temperature (B) and body weight changes (C) following intranasal SARS-CoV-2 D614G (B.1), Delta (B.1.617.2), or Omicron BA.1.1 (B.1.1.529) inoculation throughout the 14-day experimental period. Data are presented as means ± standard deviation. * p < 0.05; ** p < 0.01; *** p < 0.005; **** p < 0.0001.
Study findings
While D614G- and Delta-infected cats became weary and had increased body temperatures between days one and three post-infection (pi), Omicron-infected cats remained subclinical. Regardless of infecting VOC, the authors detected viral RNA between days one and 14 pi in nasal secretions and OPS of the infected cats.
Shedding of viral RNA in RS was lower than in nasal secretions and OPS, with Omicron-infected cats presenting lower RNA levels in feces between days 3 and 10 pi. D614G – and Delta-infected cats had high viral loads in nasal and OPS, with viral titers ranging from 2.3 to 6.3 log10 median tissue culture infectious dose (TCID50).ml-1, while Omicron-infected cats shed significantly lower viral titers. The infectious viral titers in BALF of Omicron-infected cats were also lower compared to D614G – and Delta-infected cats, i.e., in the range of 2.8 and 3.0 log10 TCID50 ml-1 on day five pi.
The sgRNA loads detected in tissues from Omicron-infected cats were markedly lower than in D614G- and Delta-infected animals. Moreover, sgRNA was detectable only in nasal turbinate and trachea from Omicron-infected cats. Conversely, the authors detected sgRNA in the nasal turbinate, tonsils, trachea, and lungs of D614G- and Delta-infected cats.
Omicron-infected cats had low SARS-CoV-2 RNA and nucleocapsid (N) protein across all the tissues tested, indicating limited tissue distribution, infection, and replication sites of Omicron BA.1.1 in cats. Histological examinations revealed that Omicron-infected cats had epithelial necrosis in the nasal cavity with variable epithelial attenuation in the upper respiratory tract. This finding reflected Omicron’s limited pathogenicity in cats.
Omicron BA.1.1-infected cats also showed increased interferon-gamma (IFN-γ) levels on days seven and 14 pi. Additionally, Regulated on Activation, Normal T Cell Expressed and Secreted (RANTES) levels were higher in Omicron-infected cats on days three and five pi. An increased anti-inflammatory IFN-γ secretion and decreased pro-inflammatory cytokine responses explain the reduced disease severity of Omicron in cats. Further studies should investigate and compare the expression of these cytokines in the most severely affected tissues of the upper and lower respiratory tract of infected cats.
Conclusions
Studies have shown low pathogenicity of the Omicron variant in murine models of SARS-CoV-2 infection. Even in humans, Omicron infections cause mild symptoms, lower viral loads, and a much-reduced risk of hospitalization compared to previous variants. Similarly, in the present study, the Omicron BA.1.1 variant showed limited pathogenicity in the study-used highly susceptible feline model of SARS-CoV-2 infection than the D614G and Delta variants. As Omicron continues to evolve into distinct sublineages, it will be essential to conduct studies to assess its biological properties in animal models.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- The Omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and Delta variants in a feline model of SARS-CoV-2 infection, Mathias Martins, Gabriela M. do Nascimento, Mohammed Nooruzzaman, Fangfeng Yuan, Chi Chen, Leonardo C. Caserta, Andrew D. Miller, Gary R. Whittaker, Ying Fang, Diego G. Diel, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.15.496220, https://www.biorxiv.org/content/10.1101/2022.06.15.496220v2
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Antibody, Anti-Inflammatory, Assay, Cell, Cell Culture, Chemokine, Coronavirus, Coronavirus Disease COVID-19, Cytokine, Cytokines, Enzyme, Gene, Heart, Hybridization, Immunoassay, Interferon, Interferon-gamma, Kidney, Liver, Lungs, Lymph Nodes, Medicine, Necrosis, Omicron, Polymerase, Polymerase Chain Reaction, Protein, Respiratory, Respiratory Disease, Reverse Transcriptase, Ribonucleic Acid, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Small Intestine, Spleen, Syndrome, Therapeutics, Tissue Culture, Tonsil, Veterinary, Virus

Written by
Neha Mathur
Neha is a digital marketing professional based in Gurugram, India. She has a Master’s degree from the University of Rajasthan with a specialization in Biotechnology in 2008. She has experience in pre-clinical research as part of her research project in The Department of Toxicology at the prestigious Central Drug Research Institute (CDRI), Lucknow, India. She also holds a certification in C++ programming.
Source: Read Full Article