The cupboard in Nicolás Rascovan’s microbial paleogenomics lab at Institut Pasteur in Paris, France, is filled up with cardboard boxes that look as if they were shipped from an office supply store. Yet, instead of pencils and Post-it notes, the boxes are filled with ancient human remains from South America — several-thousand-year-old vertebrae, petrus bones (which protect inner ear structures), and teeth ― all neatly packed in plastic bags. From these artifacts Rascovan hopes to retrieve DNA of ancient pathogens — which could help us better understand how microbes emerge and evolve and how pandemics spread. It could even, perhaps, rewrite history. “It’s a story of a continent in a closet,” Rascovan says.

Nicolás Rascovan’s in his microbial paleogenomics lab at the Institut Pasteur in Paris, France.
Over the past decade, technologic advances in DNA recovery and sequencing have made it possible for scientists such as Rascovan, an Argentinian molecular biologist, to analyze ancient specimens relatively quickly and affordably. They’ve been hunting for — and finding — DNA of centuries-old microbes in various archeological samples: from smallpox variola virus and M tuberculosis in mummified tissues, to the Black Death bacteria, Yesinia pestis , in neolithic teeth, to P falciparum preserved in historical blood stains.
The ultramodern Parisian offices of the microbial paleogenomics group, a team of five scientists led by Rascovan, clash with the logo they half-jokingly chose for themselves and plastered all over the lab’s walls: of a Jurassic Park–inspired dinosaur baring its giant, ancient teeth, made to look like an image seen under a microscope. Ancient teeth are certainly central to the group’s work, since it’s there where ancient pathogens’ DNA is most likely to be preserved — after death, teeth act like tiny, sealed-up boxes for microbes. “If you have a pathogen that is circulating in the blood, it will sometimes get into the teeth, and when you die, the DNA will stay there,” Rascovan says.
To process ancient teeth, Rascovan enters a lab clad head to toe in protective gear. That’s not so much to save himself from potentially deadly disease as to save the samples from contamination, he says. According to Sebastian Duchene Garzon, a microbiologist at the University of Melbourne, “the likelihood of ancient pathogen DNA leading to infections at present is remote, although certainly not impossible, because of how degraded the DNA usually is and because it would still need all the molecular machinery to infect a modern host.”
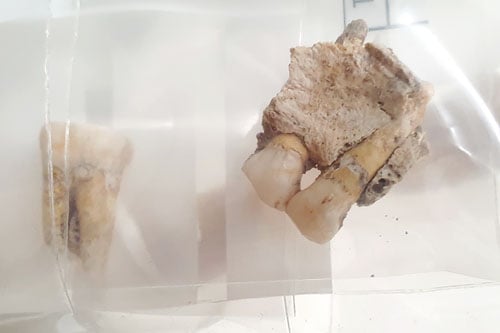
Ancient human bones excavated in South America.
To process ancient teeth in his lab, Rascovan starts with a thorough cleaning that involves bleach to remove any modern DNA contamination. Next, he cuts the tooth with a Dremel rotary tool to open it up and get into its pulp — which is not only very durable but also naturally sterile — a perfect place to find ancient pathogens. He then scrapes the pulp into a powder that can be poured into a tube for DNA extraction.
So far, Rascovan’s biggest breakthrough didn’t come from the teeth he cut up himself, though. It came from analyzing publicly available DNA data from studies of ancient human genomes. When such genomes are sequenced from fossil teeth or bones, scientists pick out the material they need for study of our ancestors’ evolutionary history. However, among the double helixes coding hominid genetic instructions often hide scraps of microbial DNA, which in the past were often simply discarded.
Rascovan downloaded data from published articles on ancient human DNA that had been found in teeth and reanalyzed them, searching for bacteria. One night, when he was alone in his office going through lines and lines of data, he spotted it: DNA of the plague-causing bacteria, Yersinia pestis. When Rascovan cross-checked to determine in which samples the bacteria’s DNA was found, his heart raced. “It was not supposed to be there,” he says. He had just discovered the most ancient case of plague in humans — which occurred 4900 years ago in Sweden.

Nicolás Rascovan holding a many-thousand-year-old human fossil.
Scientists used to believe that plague pandemics came to Europe from the Eurasian Steppe. Yet here was the DNA of Yersinia pestis lodged in the teeth of two farmers, a woman and a man, who died in Scandinavia before the plague’s supposed arrival from the East. Their bodies were buried in an unusually large common grave — of itself a possible indication of an epidemic.
When Rascovan and his colleagues applied molecular-clock analyses of the phylogenetic tree of the plague bacteria and compared various strains to see which one was the most ancestral, they confirmed that the Swedish strain of Yersinia pestis, named Gok2, was indeed the oldest — the origin of the Steppe strains rather than its distant cousin. Plague, it seemed, wasn’t brought to Europe during mass migrations from the East. Instead, it might have originated there.
Such work is not simply about rewriting history. By updating our knowledge of ancient pandemics, we can learn how different factors influence each other in fostering outbreaks. For Rascovan, the Swedish plague story underscores the importance of our lifestyle and environment for the emergence and spread of dangerous pathogens. The Gok2 strain didn’t contain a gene that makes plague particularly virulent, called ymt, yet it might have played an important role in Bronze Age Europe. At that time, mega-settlements of 10,000 to 20,000 people existed in what is now Ukraine, Romania, and Moldova, yet those settlements were frequently burned to the ground and abandoned. According to Rascovan and his colleagues, that could fit with the plague pandemic story (although this remains very much a hypothesis).
In Mexico, environmental factors might have played an important role in the severity of the 16th century “cocoliztli” epidemic (the word means “pestilence” in a local language), considered one of the most devastating epidemics in New World history. The disease, which caused vomiting, red spots on the skin, and bleeding from various body orifices, didn’t have a known cause. Some hypothesized the bug might have been smallpox, judging by the severity of the outbreak. A 2018 study of a victim’s DNA showed it contained the genome of Salmonella enterica, a bacterium that causes enteric fever — a microbe generally milder than smallpox. The study’s authors argued that specific conditions may have been necessary at the onset of the epidemic for the Salmonella enterica microbe to cause such devastating outcomes. A mix of severe draught, forced relocations of the local population by their Spanish rulers, and new subsistence farming practices all negatively affected hygienic conditions in the local settlements. According to Rascovan, such research can “place pandemics into their broader context” — with potential lessons for the future.
One of the microbes Rascovan and his team are hoping to find in the ancient teeth stocked in their lab’s closet is tuberculosis — a pathogen that kills 1.5 million people a year, yet whose evolutionary history remains largely a mystery. The focus of Rascovan and his colleagues remains on fossils shipped from South America, since we still know very little about microbes that were associated with pre-Columbian populations. South Americans have been isolated from the rest of the world for 20,000 years, making them particularly interesting candidates for the study of emergence, evolution, and spread of pathogens.
Rascovan believes that ancient microbial genomic data can help scientists better understand antibiotic resistance through comparisons of bacterial evolution before and after the discovery of antibiotics. In general, he says, by only studying current pathogens and the modern outbreaks they cause, we see only a narrow sample of something that is much more diverse and much larger. “We are missing an important part of information. Ancient samples can bring us a perspective,” he says.
For more news, follow Medscape on Facebook, Twitter, Instagram, and YouTube.
Source: Read Full Article