You’re invited to join the search for COVID-19 treatments—no lab coat or Ph.D. necessary.
Anyone with a smartphone can download the app ViDok, which lets users pick from a library of molecules that might bind to key proteins on the SARS-CoV-2 virus, which causes COVID-19, and then can tweak the molecules to try to find a better fit.
“This is a human learning process and a real experimental process,” says Thanh Truong, University of Utah professor of chemistry and designer of ViDok. “You try and you learn. And the beautiful thing is that different people look at things differently. You can improve sequentially.”
Find the ViDok app on the iOS App Store and on Google Play.
Crowdsourcing the process
The process of discovering and testing new drugs is long, on the order of years, and costly, on the order of hundreds of millions of dollars. The first steps are identifying the drug target—in this case a protein on the SARS-CoV-2 virus called a protease—and looking for molecules that can bind to that target, interrupting the virus’ reproduction cycles and effectively stopping it in its tracks. The target, Truong says, is like a lock and the process of discovering a drug is like finding just the right key.
The SARS-CoV-2 virus has only been around for less than a year, and while the race for a vaccine continues, researchers are also looking for ways to treat people already sick with COVID-19. It’s in the phase of trying out different molecules, to see if they might serve as suitable keys, that Truong has turned to the crowd for help to shorten the development time and reduce the overall costs.
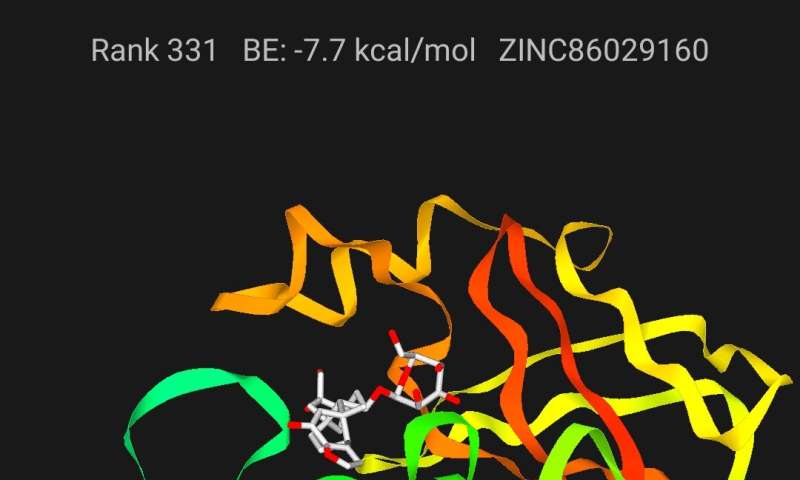
Finding new keys
Truong developed the ViDok app with the help of colleagues at the Institute for Computational Science and Technology in Vietnam. It’s designed to be easily usable by anyone, from high school chemistry students to veteran biochemistry researchers. The premise is simple: Choose a molecule, or ligand, from a library of molecules that are likely to bind well to the SARS-CoV-2 protein. Then, after determining the binding energy, or binding strength, of the molecule to the protein, the user can alter the structure of the molecule and test the binding energy again.
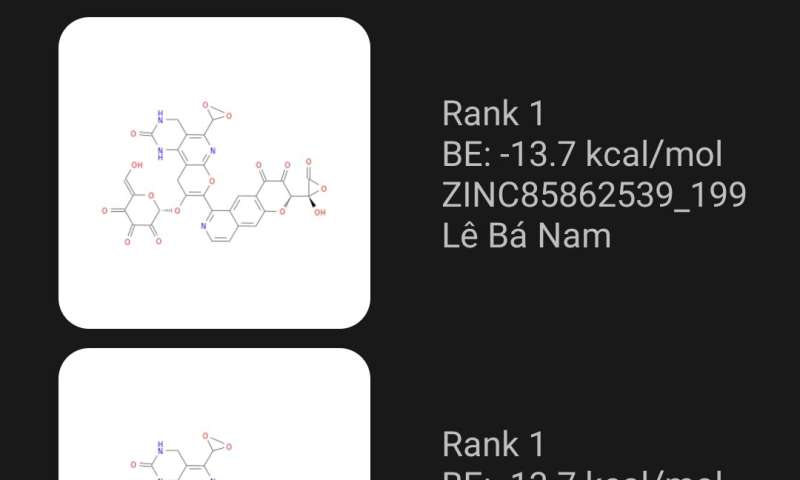
Lower binding energies are better, and after each simulation, the app will rank the new molecule against others submitted by other users. Processing of the binding energy simulations is carried out with the help of the U’s Center for High Performance Computing, which also hosts a website with all of the molecules generated by the app, with their calculated binding energies. These data are freely available for download for further research.
Truong hopes that the spirit of friendly competition can lead to advances in discovery.
“Let’s say your design ranks No. 100 and someone looks at it and says, ‘Hey, I could do better!'” he says. “Then take your results and modify it. And suddenly, he ranks No. 50. So you take his results and you modify a little bit. And you rank No. 30. This is allowing people collaborative learning. You take the results from everybody else and you build upon them.”
From app to the lab
Until now, Truong has been testing the app with a handful of people in Vietnam. Of the top 200 molecules from more than 2500 drug candidates that have been tested so far, only five of them come from the original library of potentially therapeutic molecules. The other 195 have been modified by users.
“And hopefully at the end,” Truong says, “we get a list of the best possible drug leads that experimentalists can actually go into the lab and synthesize it and then perform further studies. What users have done so far is a scientific advancement already. I can take the top results and start writing scientific papers right now. In fact, some have contacted me for the possibility of using the results for further research.”
Source: Read Full Article


