A research team led by Dr. Margaret E. Ackerman has observed elevated cross-reactive antibodies to endemic coronaviruses (CoVs), particularly to β-CoV OC43, in severe acute respiratory coronavirus 2 (SARS-CoV-2) natural infection cohorts when compared to naive controls.
Pre-existing antibodies to endemic CoVs that cross-react with SARS-CoV-2 have the potential to influence the antibody response to the SARS-CoV-2 vaccination and infection. The magnitude of cross-reactive antibodies found in the current study published on the medRxiv* preprint server correlated well with responses to the SARS-CoV-2 spike protein and the S2 subdomain.
 Study: Boosting of cross-reactive antibodies to endemic coronaviruses by SARS-CoV-2 infection but not vaccination with stabilized spike. Image Credit: P. Heitmann / Shutterstock.com
Study: Boosting of cross-reactive antibodies to endemic coronaviruses by SARS-CoV-2 infection but not vaccination with stabilized spike. Image Credit: P. Heitmann / Shutterstock.com
Does ‘original antigenic sin’ and ‘antibody-dependent enhancement’ apply to SARS-CoV-2 exposure?
The human immune system tends to preferentially utilize immunological memory based on a primary infection when a secondary and slightly different variant of that pathogen is encountered, thus resulting in a biased response to the novel strain. The phenomenon, which is otherwise known as original antigenic sin, may reduce the formation of neutralizing antibodies, thereby dampening the effective clearance of the novel virus.
This phenomenon has been established for influenza and dengue viruses and has recently been suggested to exist for SARS-CoV-2 infection. This implies that lower titers of neutralizing antibodies against SARS-CoV-2 may result from the boosting of pre-existing cross-reactive viruses.
Another phenomenon that has been proposed to exist for SARS-CoV-2 infection is an antibody-dependent enhancement (ADE). ADE is described as when cross-reactive antibodies induced by previous exposure to a related pathogen promote infection of cell types bearing antibody Fc receptors, thus potentially raising morbidity and mortality.
Though observed in vitro, there has not been abundant evidence for the existence of ADE in the context of the SARS-CoV-2 in vivo. However, the establishment of ADE in cases of dengue fever has motivated concerns about the coronavirus disease 2019 (COVID-19) enhancement.
About the study
The spike protein sequences of endemic CoVs were aligned with that of SARS-CoV-2 to compare the sequence conservation across human CoVs. Structural conservation was evaluated by superimposing SARS CoV-2 S1 and S2 domains with the most well conserved widely circulating endemic human CoV strain of OC43.
Three diverse cohorts were included in the current study to assess the antibody profiles in this observational study. The first small cohort consisted of COVID-19 convalescent individuals, for which serum and mucosal samples were available.
Secondly, a larger cohort consisted of convalescent plasma donors, acutely infected individuals, and pregnant women infected in their third trimester. The third cohort consisted of subjects for whom pre- and post-infection samples were available. A cohort of healthy adults was included that consisted of individuals vaccinated with the SARS-CoV-2 messenger ribonucleic acid (mRNA) vaccine, as well as pregnant women who were vaccinated in their third trimester. All samples were analyzed alongside samples from naive and historical negative controls.
Study findings
The N-terminal domain (NTD) of SARS-CoV-2 that contains the receptor-binding domain (RBD) showed lower sequence homology to the corresponding subdomains of endemic CoVs as compared to the S2 subdomains. However, superimposition of the SARS CoV-2 S1 and S2 domains with the most well conserved widely circulating endemic human CoV OC43 showed high structural conservation in S2 and the NTD, as well as a complete lack of homology in the RBD.
Based on both structural and sequence homology, the team suggested that preexisting antibodies raised against endemic human CoVs are more likely to target the better-conserved regions of SARS-CoV-2 like S2, and less likely to recognize the RBD.
Examination of antibody responses against endemic and pandemic human CoVs in subjects one month after SARS-CoV-2 infection found heightened OC43-specific antibody responses in SARS-CoV-2 convalescents compared to naive controls. The magnitude of these responses correlated with responses to SARS-CoV-2 spike protein and the S2 subdomain.
The team also found OC43-specific immunoglobulin G (IgG) in both the serum and mucosal samples, as well as mucosal IgA, but not IgM, in acutely infected subjects two weeks post-COVID-19 diagnosis. This finding indicates that these elevated responses were real and not due to newly induced antibody lineages with cross-reactivity to OC43. Testing of serum samples pre- and post- SARS-CoV-2 infection reinforced the hypothesis that pre-existing clonal families were boosted by the SARS-CoV-2 infection.
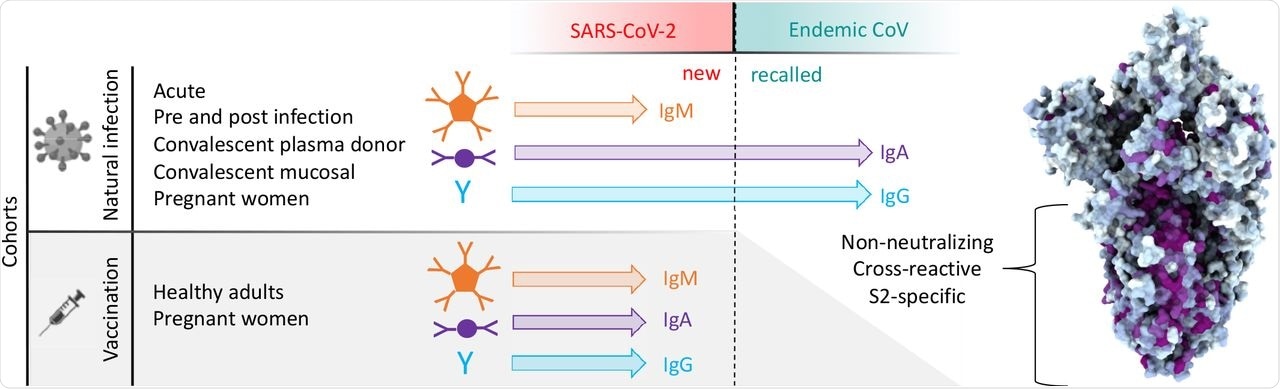 Antibody responses to SARS-CoV-2 and endemic CoV spike proteins were measured in diverse cohorts. While antibodies to SARS-CoV-2 were induced across all isotypes, only IgA and IgG responses to endemic CoV were robustly boosted, and only among naturally-infected but not vaccinated individuals. These recalled, cross-reactive responses to endemic CoV primarily recognized the better conserved S2 domain and were non-neutralizing. While other antiviral activities of broadly cross-reactive S2-specifc antibodies are not known, the differing antigenicity of natural infection and vaccination with stabilized pre-fusion spike has potential implications for the breadth and level of protection afforded by each.
Antibody responses to SARS-CoV-2 and endemic CoV spike proteins were measured in diverse cohorts. While antibodies to SARS-CoV-2 were induced across all isotypes, only IgA and IgG responses to endemic CoV were robustly boosted, and only among naturally-infected but not vaccinated individuals. These recalled, cross-reactive responses to endemic CoV primarily recognized the better conserved S2 domain and were non-neutralizing. While other antiviral activities of broadly cross-reactive S2-specifc antibodies are not known, the differing antigenicity of natural infection and vaccination with stabilized pre-fusion spike has potential implications for the breadth and level of protection afforded by each.
The SARS-CoV-2 S2 domain and whole spike (S-2P) specific antibodies isolated from convalescent serum were cross-reactive to the spike of OC43; however, RBD-specific antibodies were not. Similarly, antibodies isolated using OC43 were cross-reactive with SARS-CoV-2 S-2P and S2 but not the RBD, thereby indicating that cross-reactivity between these viral spike proteins has a link to the more conserved S2 domain. Both OC43 S-specific and SARS-CoV-2 S2-specific antibodies were non-neutralizing, which is consistent with the fact that neutralizing antibodies tend to target the RBD of SARS-CoV-2.
As compared to naturally infected and vaccinated subjects representing exposure to unstabilized and proline stabilized S antigens, respectively, the immunogens starkly contrasted. The immune response generated from vaccination with the SARS-CoV-2 spike protein stabilized in the pre-fusion conformation had either very weak or no cross-reactive profiles, thus suggesting critical importance of the conformational state of the spike protein to resulting humoral responses.
Moreover, the high levels of neutralization activity resulting from vaccination suggest favorable antigenicity of the pre-fusion conformation of the spike protein, and that the costs of original antigenic sin might be avoided by immunogen design.
“In sum, this study provides evidence that antibodies targeting OC43 are robustly boosted in response to SARS-CoV-2 infection but not vaccination with stabilized S, and that the S2 subdomain of the spike protein is likely responsible for triggering a recalled, IgG-dominated response.”
Future directions
Neutralization is not the only mechanism by which antibodies confer protection, as cross-reactive antibodies could also interact with the receptors for Fc region of antibodies found on the surface of innate immune cells and promote protective effector functions.
Therefore, the researchers of the current study emphasize the need for further work aimed at characterizing the effector function potential of these antibodies. Future work should also be focused on understanding their role in vaccine-mediated protection to provide a more complete picture of the relative risks and benefits of this recall response relevant to vaccine design, particularly in the context of viral variants of concern.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Crowley, A.R., Natarajan, H., Hederman, A. P., et al. (2021) Boosting of cross-reactive antibodies to endemic coronaviruses by SARS-CoV-2 infection but not vaccination with stabilized spike. medRxiv. doi:10.1101/2021.10.27.21265574, https://www.medrxiv.org/content/10.1101/2021.10.27.21265574v1
Posted in: Medical Science News | Medical Research News | Medical Condition News | Disease/Infection News
Tags: Antibodies, Antibody, Cell, Convalescent Plasma, Coronavirus, Coronavirus Disease COVID-19, Dengue Fever, Fever, Immune Response, Immune System, Immunoglobulin, in vitro, in vivo, Influenza, Mortality, Pandemic, Pathogen, Proline, Protein, Receptor, Research, Respiratory, Ribonucleic Acid, SARS, SARS-CoV-2, Severe Acute Respiratory, Spike Protein, Vaccine, Virus

Written by
Namita Mitra
After earning a bachelor’s degree in Veterinary Sciences and Animal Health (BVSc) in 2013, Namita went on to pursue a Master of Veterinary Microbiology from GADVASU, India. Her Master’s research on the molecular and histopathological diagnosis of avian oncogenic viruses in poultry brought her two national awards. In 2013, she was conferred a doctoral degree in Animal Biotechnology that concluded with her research findings on expression profiling of apoptosis-associated genes in canine mammary tumors. Right after her graduation, Namita worked as Assistant Professor of Animal Biotechnology and taught the courses of Animal Cell Culture, Animal Genetic Engineering, and Molecular Immunology.
Source: Read Full Article