The microscopic populations of bacteria in our intestines are, in some ways, just like us: They live in communities, eat, work, reproduce, and eventually die. Many of these bacterial species live in harmony with our bodies, providing benefits to us in exchange for nutrients and shelter. When the right kinds of bacteria cannot establish the proper symbiotic relationship with our bodies, we may be at a greater risk for a variety of immune, neurological, and metabolic diseases.
Now, in collaboration with the Broad Institute of MIT and Harvard, Caltech researchers have discovered that a certain species of bacteria behaves differently depending on where in the gut it takes up residence. The work was done in mouse models, but mapping the geography of microbial populations in the gut—collectively referred to as the gut microbiome—may be crucial for one day being able to treat and reshape a human microbiome gone awry.
The research was conducted primarily in the laboratory of Sarkis Mazmanian, Luis B. and Nelly Soux Professor of Microbiology and Heritage Medical Research Institute Investigator. A paper describing the research appears on March 9 in the journal Nature Microbiology.
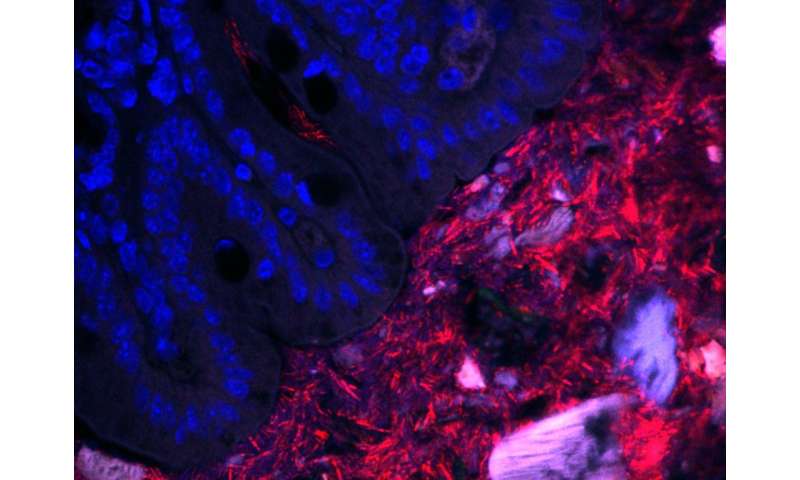
Over many years, the Mazmanian laboratory has described how Bacteroides fragilis in the gut produces beneficial molecules that protect mice from inflammatory bowel disease and autism-like symptoms. Like a densely populated city, a vast majority of the B. fragilis in the gut live within the central part of the intestinal tube, called the lumen. However, the Mazmanian laboratory discovered in 2013 that some B. fragilis reside in the bacterial equivalent of small towns, nestled into microscopic pockets within the tissue walls lining the tube. These sparse populations are protected by mucus and are largely unaffected by antibiotics, suggesting that they act as population reservoirs that ensure long-term colonization.
“For humans, where we live can dictate how we behave—for example, a person living in a city likely has a different everyday life than a person living in a small rural community,” says former graduate student Gregory Donaldson (Ph.D. ’18), the first author on the new paper. “For the bacteria that we study, the intestines represent their entire world, so we wanted to know how differently they behave depending on how far away from the intestinal surface they are.”
Though they may live in different habitats within the gut, these B. fragilis populations all have the same genetic code. What may differ, however, is how they express those genes—is a bacterium expressing a gene for replication and division, for example, or perhaps for an enzyme that digests food? Donaldson aimed to measure and compare gene expression in these two populations (intestinal wall tissue and lumen of the gut) to determine what, if any, differences were seen.
This posed a technical challenge. Because the population of bacteria living in the tissue lining is so small, their genetic material becomes obscured during sequencing by the genetic material of the mouse cells, which is far more abundant than that of the bacteria. Though mice and bacteria are distinctly different genetically, sifting through the mouse RNA to find the bacterial RNA is like finding a needle in a haystack.
Here, a crucial collaboration with Ashlee Earl of the Broad Institute made the research possible. Earl and her team led the development of a new technique, called hybrid selection RNA-sequencing, designed to fish out the elusive strands of bacterial RNA like using a magnet to search for the needles in a haystack.
“Inspired by a previous approach to sequence small populations of parasites in human blood, we developed a technique that could boost the amount of bacterial RNA we could detect in these host-rich samples by orders of magnitude,” says Earl. “This technique not only helped to reveal a new aspect of the Bacteroides-host relationship, but now provides us with a more general tool for listening in on conversations between humans and their rarest inhabitants.”
This technique revealed that B. fragilis residing in the gut tissue are surprisingly metabolically active and thriving despite their sparser population. Donaldson and his team found that one particular gene in these bacteria help them to establish a foothold in the tissue lining. Without this gene, they are unable to colonize this habitat, which is detrimental to their long-term colonization of the animal.
In contrast, B. fragilis living in the lumen are busy digesting food and nutrients, but they appear to be more focused on surviving than growing. This suggests that the epithelial surface is actually the preferred habitat for these bacteria.
As B. fragilis in the gut is known to provide beneficial protections for the host, this work suggests that a spatially intimate relationship is necessary for these positive effects. Ultimately, understanding how bacteria establish residency and persist in the gut may lead to new strategies to reinforce a healthy human microbiome to prevent or treat disease.
“While we know a great deal about which bacteria reside in humans through DNA sequencing techniques and how microbial community membership changes as a result of disease or other factors, we know far less what bacteria are doing while living inside of us,” says Mazmanian. “For the first time, this work gives us a glimpse into the lifestyle and behavior of an important human gut bacteria, while colonizing mice. Knowing what key bacteria are actually doing in the gut may help develop rationale and robust therapies from the microbiome.”
The paper is titled “Spatially distinct physiology of Bacteroides fragilis within the proximal colon of gnotobiotic mice.” In addition to Donaldson, Earl, and Mazmanian, additional co-authors are Wen-Chi Chou, Abigail Manson, Peter Rogov, James Bochicchio, Dawn Ciulla, Alexandre Melnikov, and Georgia Giannoukos of the Broad Institute of MIT and Harvard; Thomas Abeel of the Broad Institute of MIT and Harvard and Delft University of Technology; and Peter Ernst and Hiutung Chu of the UC San Diego. Funding was provided by the National Institutes of Health, the Heritage Medical Research Institute, and the National Science Foundation.
Source: Read Full Article


