Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, belongs to the Coronaviridae family. The virus consists of a single-stranded RNA genome and relies heavily on the use of the host’s cells to help it replicate and spread within the body. The pandemic has necessitated further research into how this virus works in order to fully understand and conquer this deadly disease.
Researchers in the UK, including the University of Oxford and Glasgow, as well as the University of Heidelberg, published their analysis of protein-RNA interactions in SARS-CoV-2 and the subsequent significance in the journal Molecular Cell. This research could help to uncover novel antiviral strategies for COVID-19 due to a broader understanding of how the virus works.
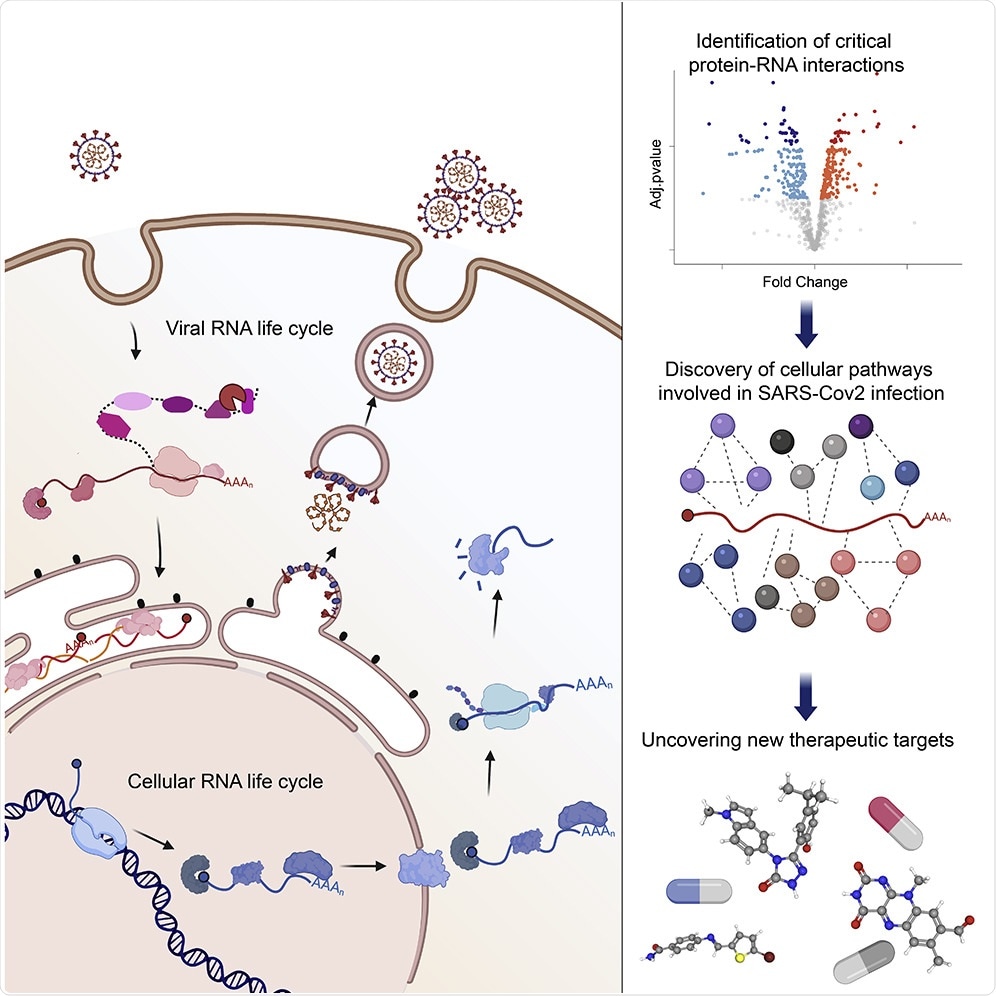
Understanding the Life Cycle
RNA viruses have life cycles that predominantly rely on the host’s resources, with most of its processes consisting of finding a path to multiply, transport, and deliver the viral RNA genome into a new cell. However, being prevented by the virus’s own inadequacies of being unable to encode the proteins that may be required to achieve the objectives, the viruses hijack and utilize the cellular RNA-binding proteins (RBPs).
The virus responsible for COVID-19 relies on cellular RBPs in order to replicate and increase its numbers within the body. However, it is not fully known which RBPs control the virus’ life cycle, which was the main aim of the researchers, in order to characterize the cellular and viral RBPs involved in COVID-19 infections.
These RBPs can either work to help the virus replicate or trigger the cellular antiviral state, suppressing viral gene expression; it can work to either promote or restrict infections, and this makes RBPs a vital regulator of the virus’ life cycle.
Global Analysis
The researchers found that the COVID-19 virus remodels the cellular RNA-bound proteome. This can cause wide-reaching impacts on other components such as RNA metabolic pathways, non-canonical RBPs and antiviral factors.
A new method was also used in this global analysis in order to identify the proteins that have direct interaction with the viral RNA, which resulted in the findings of previously obscure cellular RBPs as well as six viral proteins.
The analysis undertaken by these postdoctoral researchers also illustrates how the SARS-CoV-2 infection is regulated by components of a transfer RNA (tRNA) complex, as well as the drugs available which can inhibit the infection through targeting the host RBPs that interact with the virus. This is significant as through directing the strategy towards the RBPs that are within the host, it is possible to decrease or even prevent the interaction the RBPs have with the virus, which could hopefully inhibit the replication of the virus within the body, inhibiting the infection.
Therapeutic Significance
The importance of identifying and understanding the role of key regulators lies in their use in being a target for therapeutics. With many potential therapeutic targets found, such as critical cellular proteins as well as their drug-targeting counterparts, “it will be possible to identify novel antivirals,” commented Alfredo Castello, a researcher who helped lead the study. He states that their “efforts, together with those of the scientific community, should focus now on testing these drugs in infected cells and animal models to uncover which ones are the best antivirals.”
Professor Shabaz Mohammed also includes the interaction and partnership between the Castello and Mohammed labs using the Sindbis virus as a discovery model that helped discover viral RNA interactors, enabling this team to work fast at the start of the pandemic. He remarked that their “methodology will now be ready to respond rapidly to future viral threats.”
The collaborative efforts of these researchers within the global analysis of protein-RNA interactions in SARS-CoV-2 infected cells enabled key regulators of the infection to be uncovered, as well as providing insight into the available drugs which target these. This will hopefully enable novel therapeutics to be developed, which can effectively prevent viral replication of SARS-CoV-2 as well as other potentially dangerous viruses, especially in the midst of new variants being found.
- Kamel, W., Noerenberg, M., Cerikan, B., Chen, H., Järvelin, A.I., Kammoun, M., Lee, J.Y., Shuai, N., Garcia-Moreno, M., Andrejeva, A., Deery, M.J., Johnson, N., Neufeldt, C.J., Cortese, M., Knight, M.L., Lilley, K.S., Martinez, J., Davis, I., Bartenschlager, R., Mohammed, S., Castello, A., Global analysis of protein-RNA interactions in SARS-CoV-2 infected cells reveals key regulators of infection, Molecular Cell (2021), DOI: https://doi.org/10.1016/j.molcel.2021.05.023, https://www.sciencedirect.com/science/article/pii/S109727652100407X?via%3Dihub
Posted in: Medical Research News | Disease/Infection News
Tags: Cell, Coronavirus, Coronavirus Disease COVID-19, Drugs, Gene, Gene Expression, Genome, Pandemic, Protein, Proteome, Research, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Therapeutics, Transfer RNA, Virus

Written by
Marzia Khan
Marzia Khan is a lover of scientific research and innovation. She immerses herself in literature and novel therapeutics which she does through her position on the Royal Free Ethical Review Board. Marzia has a MSc in Nanotechnology and Regenerative Medicine as well as a BSc in Biomedical Sciences. She is currently working in the NHS and is engaging in a scientific innovation program.
Source: Read Full Article


