Researchers and data scientists at UT Southwestern Medical Center and MD Anderson Cancer Center have developed an artificial intelligence technique that can identify which cell surface peptides produced by cancer cells called neoantigens are recognized by the immune system.
The pMTnet technique, detailed online in Nature Machine Intelligence, could lead to new ways to predict cancer prognosis and potential responsiveness to immunotherapies.
“Determining which neoantigens bind to T cell receptors and which don’t has seemed like an impossible feat. But with machine learning, we’re making progress,” said senior author Dr. Tao Wang, Ph.D., Assistant Professor of Population and Data Sciences, and with the Harold C. Simmons Comprehensive Cancer Center and the Center for Genetics of Host Defense at UT Southwestern.
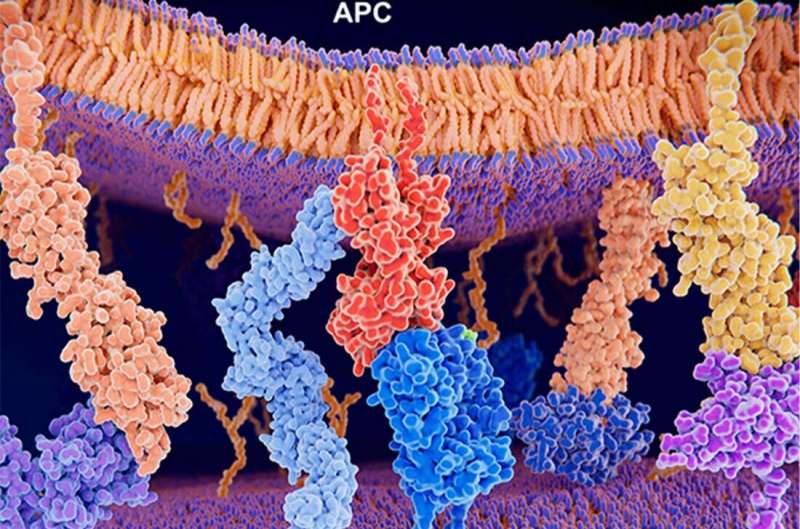
Mutations in the genome of cancer cells cause them to display different neoantigens on their surfaces. Some of these neoantigens are recognized by immune T cells that hunt for signs of cancer and foreign invaders, allowing cancer cells to be destroyed by the immune system. However, others seem invisible to T cells, allowing cancers to grow unchecked.
“For the immune system, the presence of neoantigens is one of the biggest differences between normal and tumor cells,” said Tianshi Lu, first co-author with Ze Zhang, doctoral students in the Tao Wang lab, which uses state-of-the-art bioinformatics and biostatistics approaches to study the implications of tumor immunology for tumorigenesis, metastasis, prognosis, and treatment response in a variety of cancers. “If we can figure out which neoantigens stimulate an immune response, then we may be able to use this knowledge in a variety of different ways to fight cancer,” Ms. Lu said.
Being able to predict which neoantigens are recognized by T cells could help researchers develop personalized cancer vaccines, engineer better T cell-based therapies, or predict how well patients might respond to other types of immunotherapies. But there are tens of thousands of different neoantigens, and methods to predict which ones trigger a T cell response have proven to be time-consuming, technically challenging, and costly.
Searching for a better technique with support of grants from the National Institutes of Health (NIH) and Cancer Prevention and Research Institute of Texas (CPRIT), the research team looked to machine learning. They trained a deep learning-based algorithm that they named pMTnet using data from known binding or nonbinding combinations of three different components: neoantigens; proteins called major histocompatibility complexes (MHCs) that present neoantigens on cancer cell surfaces; and the T cell receptors (TCRs) responsible for recognizing the neoantigen-MHC complexes. They then tested the algorithm against a dataset developed from 30 different studies that had experimentally identified binding or nonbinding neoantigen T cell-receptor pairs. This experiment showed that the new algorithms had a high level of accuracy.
The researchers used this new tool to gather insights on neoantigens cataloged in The Cancer Genome Atlas, a public database that holds information from more than 11,000 primary tumors. pMTnet showed that neoantigens generally trigger a stronger immune response compared with tumor-associated antigens. It also predicted which patients had better responses to immune checkpoint blockade therapies and had better overall survival rates.
Source: Read Full Article


