The development and emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern that could potentially evade a vaccine-induced immune response is an ongoing concern, with the alpha (B.1.1.7), beta (B.1.351), gamma (B.1.248), and delta (B.1.614.2) lineages having largely independently evolved mutations to the spike protein and receptor binding domain that enhance transmissibility and in some cases provide mild resistance to neutralizing antibodies.
In a research paper recently uploaded to the preprint server bioRxiv* by Tada et al. (July 19th, 2021) the neutralizing capacity of antibodies generated by either messenger RNA (mRNA) or adenoviral vector-based vaccines against Beta, Delta, Delta plus, and Lambda variants are investigated, among others, highlighting the importance of continued SARS-CoV-2 genomic surveillance.
.jpg)
How was the study performed?
The Pfizer-BioNTech, Moderna, and Johnson & Johnson vaccines have each been approved for emergency use by the U.S. Food and Drug Administration (FDA), with the former two being mRNA-based while the latter utilizes an adenovirus vector.
Sera were collected from individuals having received any one of these vaccines at several time points, extending from one week to three months post-vaccination. In addition, the neutralizing activity of the generated antibodies towards several SARS-CoV-2 variants was assessed.
The key mutations that characterize each of the examined lineages were identified for better representation by the pseudoviral particles utilized, expressing the unique spike protein on their surface whose structure will determine the affinity of neutralizing antibodies towards them.
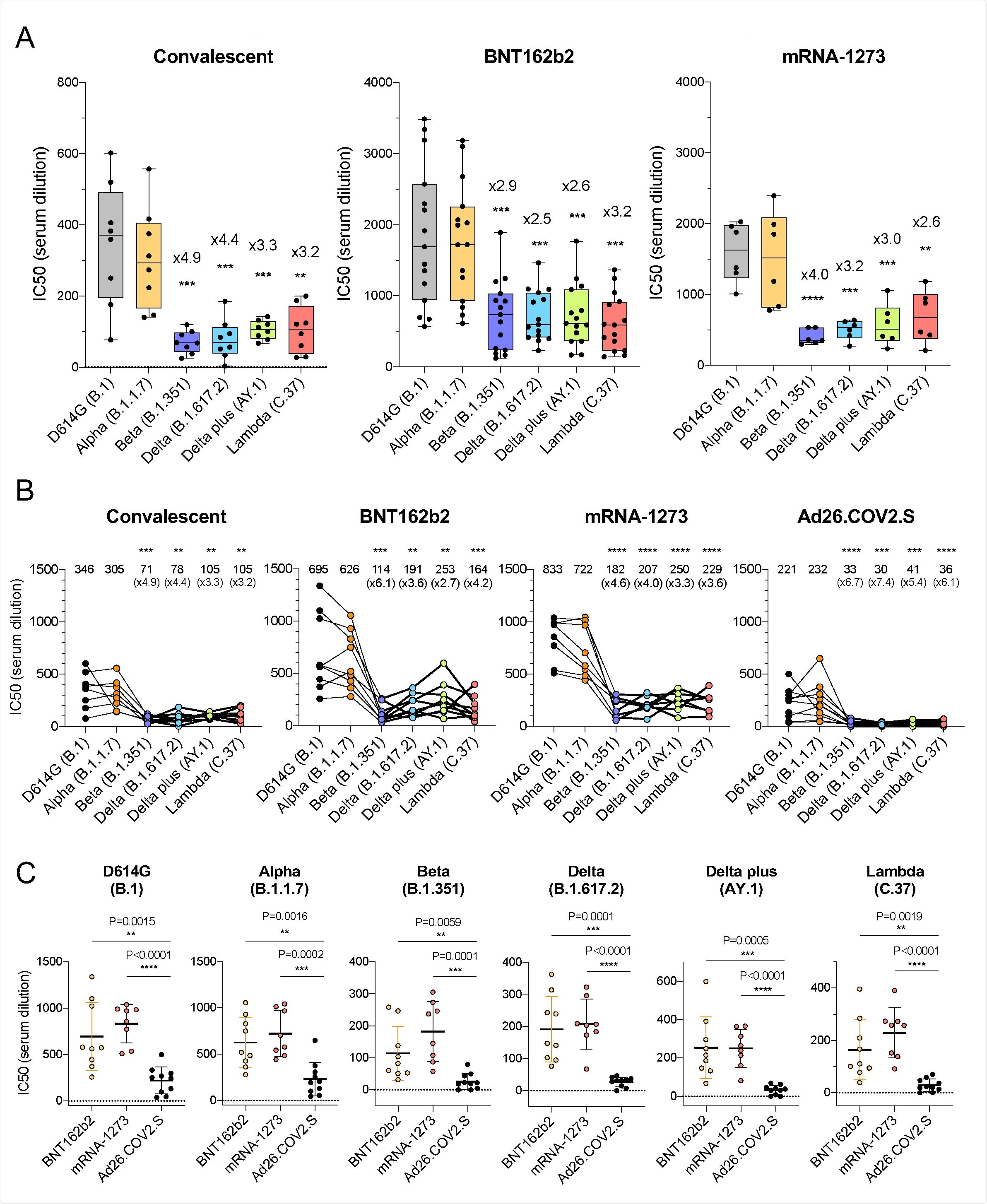
Sera from individuals that had been infected with SARS-CoV-2 prior to the emergence of these variants of concern was firstly administered, with the spike protein bearing the D614G mutation common to these variants being neutralized by the sera with antibody titers of 346 at IC50.
Comparatively, titers of only 305, 71, 78, 105, and 105 were seen upon exposure to alpha, beta, delta, delta plus, and lambda variants, respectively, indicating modest resistance to neutralization in these latter strains.
Comparison of vaccines
Sera collected from individuals having received either of the mRNA vaccines was notably more effective two weeks post-administration against the D614G strain than convalescent sera, with a subsequent and modest 2.5-4.0-fold drop in antibody titer observed towards the SARS-CoV variants of concern in comparison with D614G.
Resistance towards antibodies observed in beta, delta, and lambda variants were attributed to E484K, L452R, and L452Q and F490S mutations, respective to the strain.
Upon further investigation, the group identified the L452R/Q mutation of the lambda variant as a key factor in reduced neutralizing antibody potency, seeing a two-fold increase in viral infectivity in immunized hosts and a three-fold increase in relative affinity towards the ACE2 receptor.
Alternatively, sera collected from individuals having received the adenovirus vector-based vaccine was seen to experience an even greater drop in efficacy against the variants of concern compared to the mRNA vaccines, around 5-7 fold against the delta plus or beta variants compared to D614G. REGN10933 and REGN10987 monoclonal antibodies were also exposed to the multiple SARS-CoV-2 lineages, showing reduced activity towards the beta and delta plus variants compared with D614G, attributed to K417N and E484K mutations.
This work has demonstrated that the SARS-CoV-2 variants of concern show a degree of resistance towards convalescent or vaccine-induced antibodies. However, that the mRNA vaccines, in particular, are still likely to generate a sufficient response against any lineage three months post-administration.
The adenovirus vector-based vaccine demonstrated a more notable loss in efficacy towards the newly emerging SARS-CoV-2 variants, being able to generate a less robust antibody response. However, the authors note that while other studies have observed a slight drop in the generated antibody response by the J&J vaccine against the latter variants, the results have not been as stark as reported here, which could be a factor of demographics or some other unaccounted influence.
In any case, constant genomic surveillance and testing of SARS-CoV-2 variants of concern will be required for the foreseeable future, potentially facilitating the development of vaccine boosters that maintain the efficacy of newly emerging variants that would otherwise escape established immunity.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Comparison of Neutralizing Antibody Titers Elicited by mRNA and Adenoviral Vector Vaccine against SARS-CoV-2 Variants Takuya Tada, Hao Zhou, Marie I. Samanovic, Belinda M. Dcosta, Amber Cornelius, Mark J. Mulligan, Nathaniel R. Landau, bioRxiv, 2021.07.19.452771; doi: https://doi.org/10.1101/2021.07.19.452771, https://www.biorxiv.org/content/10.1101/2021.07.19.452771v2
Posted in: Medical Research News | Disease/Infection News | Pharmaceutical News
Tags: ACE2, Adenovirus, Antibodies, Antibody, Coronavirus, Coronavirus Disease COVID-19, Efficacy, Genomic, Immune Response, immunity, Immunization, Mutation, Protein, Receptor, Research, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Vaccine

Written by
Michael Greenwood
Michael graduated from Manchester Metropolitan University with a B.Sc. in Chemistry in 2014, where he majored in organic, inorganic, physical and analytical chemistry. He is currently completing a Ph.D. on the design and production of gold nanoparticles able to act as multimodal anticancer agents, being both drug delivery platforms and radiation dose enhancers.
Source: Read Full Article


