The Omicron variant of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported as a variant of concern (VOC) in November 2021 due to its higher transmissibility and possibly higher immune evasion capabilities. The SARS-CoV-2 Omicron variant emerged against the background of the circulating Delta SARS-CoV-2 variant; however, it rapidly outcompeted the Delta variant to become the dominant circulating strain in most of the world within a very short period.
A new study published on the preprint server medRxiv* shows that during this period, both the Omicron and Delta variants were able to infect patients with healthy immune function.
Study: Evidence of Co-Infection During Delta and Omicron Variants of Concern Co-Circulation, Weeks 49-2021 To 02-2022, France. Image Credit: Corona Borealis Studio / Shutterstock.com
Introduction
The detection of the SARS-CoV-2 ribonucleic acid (RNA) genome is achieved through the use of reverse transcriptase-polymerase chain reaction (RT-PCR) assay in samples from the respiratory tract. This test is meant to detect three viral genes including the open reading frame 1 (ORF1), nucleocapsid (N), and spike (S) genes.
When the S gene is not detected but the other two are present, it is an S gene target failure (SGTF). Thus, the detection of SGTF has been used to screen for the presence of either the SARS-CoV-2 Alpha or Omicron variants, which have a Δ69-70 deletion in the S protein, according to the national policy in France at that time. Additional mutational screening was done for the single nucleotide polymorphisms (SNPs) S:L452R and S:K417N to rapidly identify the Delta and Omicron strains, respectively.
The Omicron variant was first detected in France in December 2021 during week 49 of that year. Between then and week two of 2022, the same center had over 2,800 positive samples, of which 85% were screened for VOC mutations.
In 0.2% of these samples, both the aforementioned mutations that are characteristic of both Delta and Omicron were found, thereby indicating that both strains were coinfecting the same individual, but without the SGTF typical of the 21K Omicron variant. These patient samples were subjected to further study.
Study findings
The seven patients who tested positive for both Delta and Omicron infections were immunocompetent adults, only one of whom was over the age of 70, while the others were below the age of 35 years. All had received two or three doses of a COVID-19 vaccine and were asymptomatic or had mild symptoms, except the elderly patient who was unvaccinated and in respiratory failure at nine days post-diagnosis.
Whole-genome sequencing (WGS) was carried out on these patients. The presence of characteristic clade-defining mutations for Delta and Omicron were searched for and the proportion of each was determined. More specifically, the researchers were interested in identifying mutations at position 23604 or S:P681, where Delta and Omicron show 23604G, S:P681R, and 23604A; S:P681H, respectively, and the clade-defining mutations.
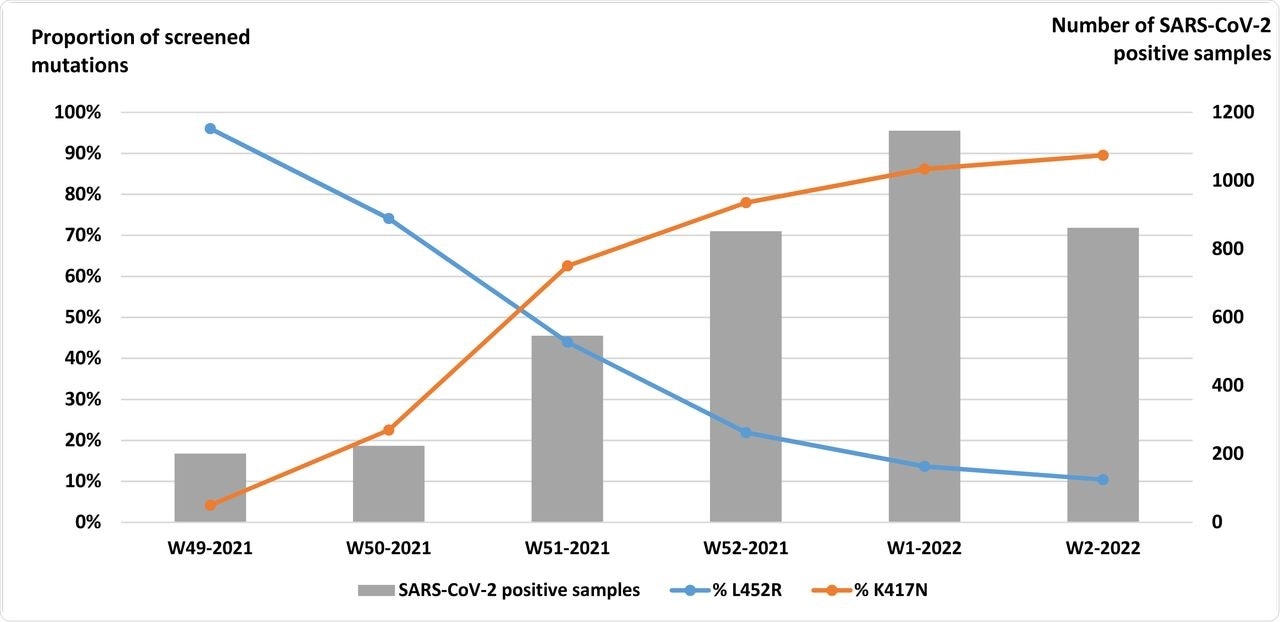
Weekly distribution of S:L452R and S:K417N single nucleotide polymorphism detection on 3,237 SARS-CoV-2 positive screened samples from weeks 49-2021 to 02-2022, University Hospital of Clermont-Ferrand, France.
Two patients were monitored with samples collected at seven and 11 days, respectively. In the first patient, the day one sample showed equal proportions of Delta and Omicron. Conversely, in the day 11 sample, the Delta variant accounted for over 96% of the sample, thus demonstrating that the Delta variant outcompeted Omicron by day 11.
For the other patient, at day one, the Delta variant made up 80% of the sample; however, at day seven, 100% of the viral population was comprised of the Omicron. In both cases, the number and percentage of defining mutations remained intact throughout the genome, ruling out any recombination between the strains.
Implications
The results show that two SARS-CoV-2 VOCs can occur when both are present in the same region and have high infectiousness. While these events are rarely reported, they may not be as uncommon as thought, since VOCs are not routinely screened for in many countries.
In contrast, the scientists in this study executed a strategy to detect coinfection.
The use of specific RT-PCR assays targeting SNP is an efficient strategy for rapid detection of circulating SARS-CoV-2 VOC and can detect co-infection only if SNPs of different variants are simultaneously screened and if the allele discrimination plots are carefully analysed.”
For instance, when additional mutations are found with the assigned clade, this could indicate the need for further analysis of WGS data. In this study, the researchers monitored low-frequency variants for clade-defining mutations and confirmed the presence of coinfections.
Interestingly, the clinical course was unaltered by the coinfection. However, the genomic data shows that each of these variants can displace the other, a finding previously reported in immunocompromised individuals. Immunologic studies could help understand the extent to which this is affected by vaccine-induced antibodies.
Despite any evidence of recombination in the two patients who were closely monitored after their initial diagnosis, there was a suggestion of this phenomenon in another patient sample, which will need further analysis. Recombination is potentially serious, as it may lead to the emergence of a new variant with higher pathogenicity or immune evasion, or transmissibility characteristics.
This underlines the need for continuous efforts to maintain an effective genomic surveillance of SARS-CoV2 infections and a re-enforced surveillance of co-infection with emerging variants.”
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information
- Combes, P., Bisseux, M., Bal, A., et al. (2022). Evidence of Co-Infection During Delta and Omicron Variants of Concern Co-Circulation, Weeks 49-2021 To 02-2022, France. medRxiv. doi:10.1101/2022.03.02.22271694. https://www.medrxiv.org/content/10.1101/2022.03.02.22271694v1.
Posted in: Genomics | Medical Research News | Disease/Infection News
Tags: Allele, Antibodies, Assay, Coronavirus, Coronavirus Disease COVID-19, covid-19, Frequency, Gene, Genes, Genome, Genomic, Hospital, Nucleotide, Omicron, Polymerase, Polymerase Chain Reaction, Protein, Respiratory, Reverse Transcriptase, Ribonucleic Acid, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Single Nucleotide Polymorphism, Single Nucleotide Polymorphisms, Syndrome, Vaccine

Written by
Dr. Liji Thomas
Dr. Liji Thomas is an OB-GYN, who graduated from the Government Medical College, University of Calicut, Kerala, in 2001. Liji practiced as a full-time consultant in obstetrics/gynecology in a private hospital for a few years following her graduation. She has counseled hundreds of patients facing issues from pregnancy-related problems and infertility, and has been in charge of over 2,000 deliveries, striving always to achieve a normal delivery rather than operative.
Source: Read Full Article


