A group of researchers at Université de Montréal has simulated the effects of more than 17,000 possible mutations of the spike protein of SARS-CoV-2, the virus responsible for COVID-19.
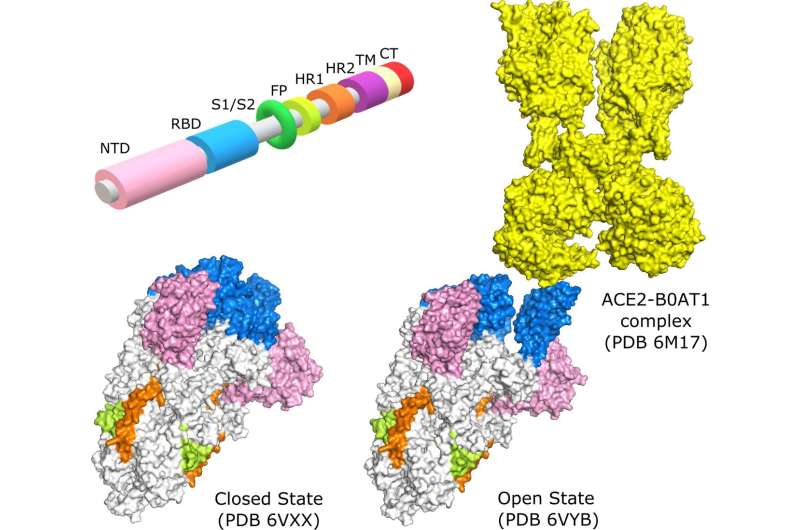
These spike proteins are found on the surface of the virus and are responsible for its toothed appearance. When these proteins are in the “open” state, the virus is able to bind to human cells and trigger an infection.
As Najmanovich, project lead and professor in UdeM’s Department of Pharmacology and Physiology, explains, “our computer modeling has shown that in the original virus the spike proteins are in the closed state 75% of the time and in the open state 25% of the time. In D614G, the very first spike mutation, the open state is more rigid and the closed state more flexible. This causes the spike proteins to stay in the open state longer, giving the virus more opportunity to bind to human receptors and making it more infectious.”
This modeling technique has enabled the team to predict future mutations even before they appear in nature and to anticipate their virulence for humans.
A proven technique, but one with limitations
Prof. Najmanovich, who is a specialist in molecular design and computational structural pharmacology, points out that this model correctly foresaw the emergence of specific mutations that appeared in several variants of concern, including B117 (the “British” variant), B1351 (the “South African” variant), and P1 (the “Brazilian” variant).
“Proteins are linear chains of 20 different types of amino acids,” explains Najmanovich. “The spike protein contains more than 1,000 amino acids. In our supercomputer simulations, we replaced each amino acid with each of the other 19 possible amino acids, and then repeated the process for each site on the original virus. This gave us not only all existing mutations, but also ones that are not yet present in nature.”
Source: Read Full Article


