In a recent study posted to the bioRxiv* pre-print server, a team of researchers demonstrated that ubiquitin-specific peptidase 22 (USP22) is a crucial regulator of basal interferon (IFN) signaling in native human intestinal epithelial cells (hIECs).
Innate immunity components, such as pattern recognition receptors (PRRs) and interferon (IFN) signaling, are closely regulated by deubiquitinating enzymes (DUBs), like USP22, that take over the host ubiquitin machinery. The cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes protein (STING) is a PPR and a sensor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection that mediates the secretion of pro-inflammatory cytokines during the infection. Upon activation, STING activates interferon-stimulated genes (ISGs) and IFNs, leading to the initiation of antiviral and inflammatory transcriptional programs.
Amid the coronavirus disease 2019 (COVID-19) pandemic, it is of utmost relevance to identify host factors that control SARS-CoV-2 infection. Although USP22 has been associated with IFN signaling and ISG expression upon viral infections, the mechanisms underlying USP22 priming of PRRs and IFN signaling in native, uninfected environments remain unknown.
Study: USP22 controls type III interferon signaling and SARS-CoV-2 infection through activation of STING. Image Credit: Juan Gaertner / Shutterstock
About the study
In the present study, researchers studied how the prolonged expression of low basal levels of type I and III IFNs primes host responses against SARS-CoV-2 in hIECs. It is worth noting here that the expression of IFN-specific receptors is restricted by cell type and determines IFN responses.
The researchers performed gene-set enrichment analysis on USP22-regulated genes to investigate if gene ontology (GO) sets specifically regulate USP22. They used quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) to validate the USP22-regulated changes in gene expression.
Wild-type (WT), non-human target cells (NHT) CRISPR/Cas9 control Caco-2 cells and USP22 knockout (KO) Caco-2 cells were subjected to infection with SARS-CoV-2 particles at a multiplicity of infection (MOI) of 1 to test the functional relevance of the increased antiviral signaling upon loss of USP22 expression. Infected Caco-2 cells, fixed at 24 hours post-infection (hpi), were subjected to quantification of SARS-CoV-2 replication via immunofluorescence to recognize SARS-CoV-2 nucleocapsid protein.
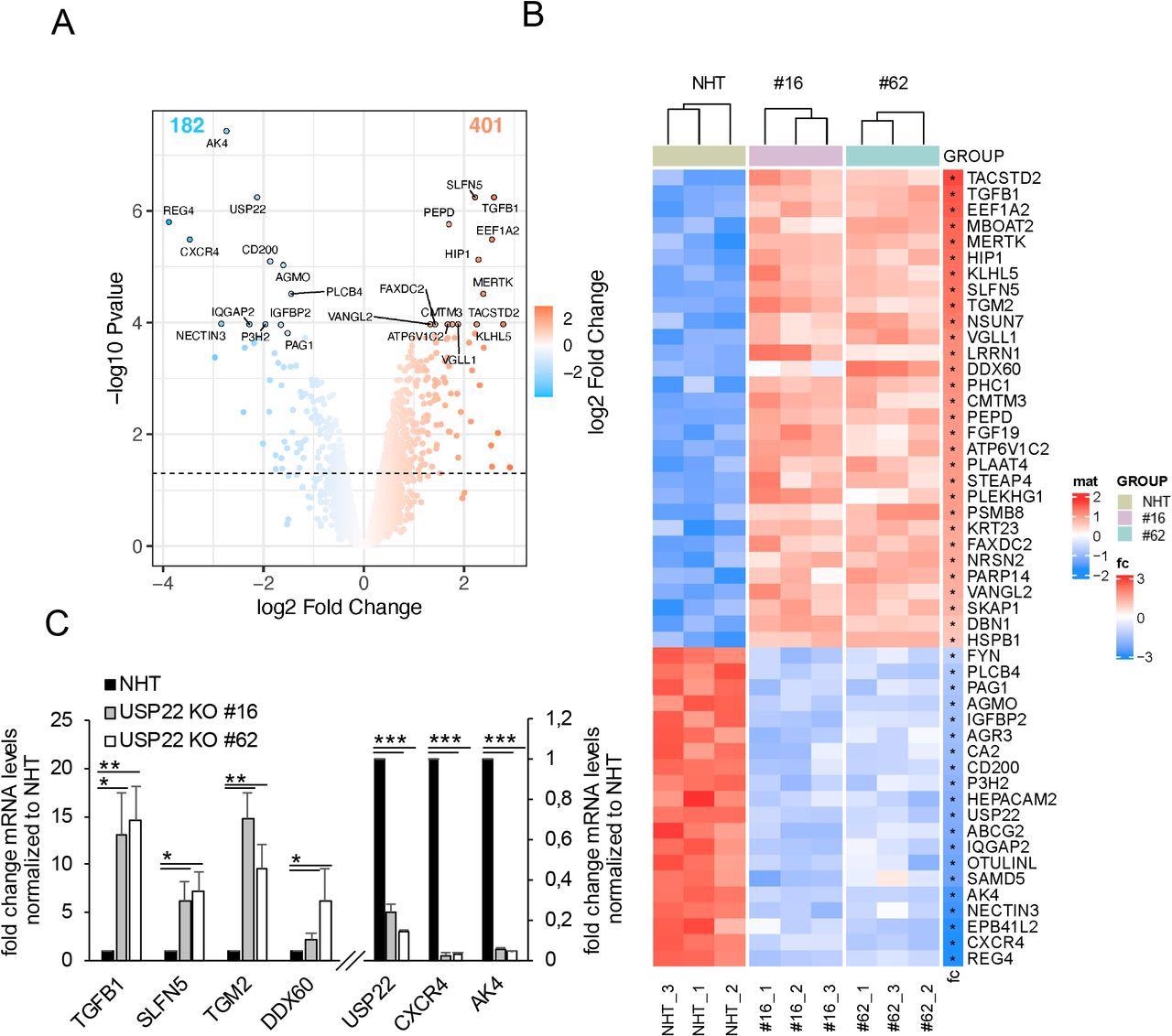
Profiling USP22-mediated gene expression in HT-29 hIECs. A. Volcano plot showing the differential gene expression patterns of two independent single-cell HT-29 USP22 CRISPR/Cas9 KO clones (#16 and #62) compared to CRISPR/Cas9 control (NHT) HT-29 cells. Color code represents the log2 foldchange compared to NHT. B. Heatmap of the top-50 differentially regulated genes between HT-29 USP22 KO single clones #16 and #62 and the NHT control. Color coding represents the row-wise scaled (Z-score) RNA intensities. Genes are sorted according to their log2 fold change, compared to NHT. C. Basal mRNA expression levels of the indicated genes were determined in control and two independent USP22 KO HT-29 single clones using qRT-PCR. Gene expression was normalized against 28S mRNA and is presented as x-fold mRNA expression compared to NHT. Mean and SD of three independent experiments in triplicate are shown. *P < 0.05; **P < 0.01, ***P < 0.001.
Study findings
USP22-mediated gene expression profiling in hIECs (cell line HT-29) identified transcriptional and extranuclear targets for USP22. The quantification of alterations in gene expression in two independent HT-29 USP22 KO single-cell clones revealed a marked alteration in gene expression, with 401 genes up-regulated and 182 down-regulated.
The differential regulation of gene expression, as well as loss of USP22 expression, was also demonstrated by qRT-PCR of the USP22-dependent up and down-regulated genes, confirming the quality of the microarray.
GO analysis revealed an enrichment of genes linked to type I and II IFN signaling. Compared to control NHT HT-29 cells, in KO HT-29 cells, USP22 regulated viral genome replication and several other viral processes, such as the IFN-γ, and IFN-γ mediated signaling pathways. The GO terms of strongly downregulated genes were enriched in mitochondrial translation and gene expression. In addition, the GO analysis showed processing of transfer ribonucleic acid (tRNA), ribosomal RNA (rRNA), and non-coding RNA (ncRNA).
qRT-PCR confirmed that the expression of several ISGs increased in two independent HT-29 USP22 KO clones, including BST2, PARP9, USP18, OAS3, IFIT1, IRF9, ISG15, OAS2, IFI27, and IFI6. In addition, loss of USP22 increased protein expression of MX1, IRF9, ISG56, and ISG20, suggesting that USP22 controls the expression of IFN signaling and viral defense genes, even in the absence of exogenous IFN stimulation or virus infection.
The immunofluorescence results showed that USP22-deficient cells displayed a substantial decrease in SARS-CoV-2 infection compared to infected control cells. Notably, USP22-deficient Caco-2 cells also expressed higher levels of IFN-λ1, compared to control cells, whereas their IFN-α and IFN-β expression remained unaltered. Together, these findings indicated that USP22 controls SARS-CoV-2 infection, replication, and the generation of de novo infectious SARS-CoV-2 particles, partially through STING, a mediator of IFN-λ1 production in HT-29 cells.
Conclusions
Cellular functions of USP22 in viral infections, interferon signaling, USP22 interactions, and USP22-mediated ubiquitination have been studied previously. According to the authors, the current study is the first study to depict the basal functions of USP22 in the regulation of ISG expression and STAT signaling in native hIECs.
These findings have important implications for SARS-CoV-2 infections. They identified USP22 as a negative regulator of type III IFN secretion in basal settings in the absence of exogenous IFNs or viral infection. In addition, it revealed that USP22 regulated both basal and 2’3’-cGAMP induced STING ubiquitination and activation. However, it remains undetermined why USP22 alters a significant fraction of IFN-unrelated genes while the expression of other genes is not altered upon the loss of USP22 expression.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- USP22 controls type III interferon signaling and SARS-CoV-2 infection through activation of STING, Rebekka Karlowitz, Megan, L. Stanifer, Jens Roedig, Geoffroy Andrieux, Denisa Bojkova, Sonja Smith, Lisa Kowald, Ralf Schubert, Melanie Boerries, Jindrich Cinatl Jr., Steeve Boulant, Sjoerd J. L. van Wijk, bioRxiv 2022.02.01.478628; doi: https://doi.org/10.1101/2022.02.01.478628, https://www.biorxiv.org/content/10.1101/2022.02.01.478628v1
Posted in: Molecular & Structural Biology | Medical Research News | Disease/Infection News
Tags: Cas9, Cell, Cell Line, Coronavirus, Coronavirus Disease COVID-19, covid-19, CRISPR, Cytokines, Gene, Gene Expression, Genes, Genome, immunity, Interferon, Knockout, Microarray, Pandemic, Polymerase, Polymerase Chain Reaction, Protein, Protein Expression, Respiratory, Reverse Transcriptase, Ribonucleic Acid, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Translation, Ubiquitin, Virus

Written by
Neha Mathur
Neha Mathur has a Master’s degree in Biotechnology and extensive experience in digital marketing. She is passionate about reading and music. When she is not working, Neha likes to cook and travel.
Source: Read Full Article