Scientists from Israel have recently estimated the levels of circulating microRNAs (miRNAs) in hospitalized coronavirus disease 2019 (COVID-19) patients with the aim of identifying prognostic biomarkers for disease severity. They have identified miR-155, an important regulatory molecule in the immune system, as a potential biomarker to predict COVID-19 severity and mortality. The study is published in the Journal of Personalized Medicine.
 Study: miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients. Image Credit: ART-ur / Shutterstock
Study: miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients. Image Credit: ART-ur / Shutterstock
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of COVID-19, has been found to cause a range of clinical consequences in affected individuals. While mild symptoms have been observed in about 80% of infected individuals, about 20% of patients have been found to develop severe and often fatal COVID-19. Therefore, to effectively manage the disease condition and reduce associated morbidity and mortality, it is vital to identify potent prognostic biomarkers that accurately distinguish highly susceptible individuals from less susceptible individuals.
The microRNAs (miRNAs) are short stretches of nucleotides that negatively regulate the expression of mRNAs at the translational level. Circulating miRNAs that are secreted by damaged or apoptotic cells act as intercellular signaling molecules to regulate the activity of recipient cells. In recent years, circulating miRNAs have shown promising outcomes in predicting the clinical course of many disease conditions.
In the current study, the scientists have assessed the levels of circulating miRNAs including miR-21, miR-146a, miR-146b, miR-155, and miR-499 in hospitalized COVID-19 patients. Given their association with COVID-19-related molecular pathways, the scientists have selected these miRNAs and investigated their ability to predict the clinical severity of COVID-19.
Study design
The circulating miRNA levels were measured in 37 adult COVID-19 patients and compared with that in 15 healthy adults. In the COVID-19 cohort, both hospitalized patients with mild infection (no respiratory support or oxygen supplementation) and severe infection (requiring invasive or non-invasive ventilation) were included.
Blood samples collected from the patients and healthy individuals were processed for measuring the levels of circulating miRNAs.
Expression of miRNAs in hospitalized COVID-19 patients
The study identified two miRNAs, namely miR-155 and miR-146b, that showed significantly different expression profiles between COVID-19 patients and healthy individuals. Specifically, a relatively lower expression of miR-155 was observed in COVID-19 patients compared to that in healthy individuals. In contrast, the exact opposite expression profile was observed for miR-146b.
Among two identified miRNAs, miR-155 showed significantly different expression profiles between mild patients, severe patients, and healthy individuals. Compared to healthy individuals, mild and severe COVID-19 patients showed 2.5-fold and 5-fold lower circulating miR-155, respectively.
In the case of miR-146b, a relatively less pronounced difference in expression was observed. Compared to healthy individuals, mild and severe COVID-19 patients showed 2.3-fold and 2-fold higher levels of circulating miR-155, respectively.
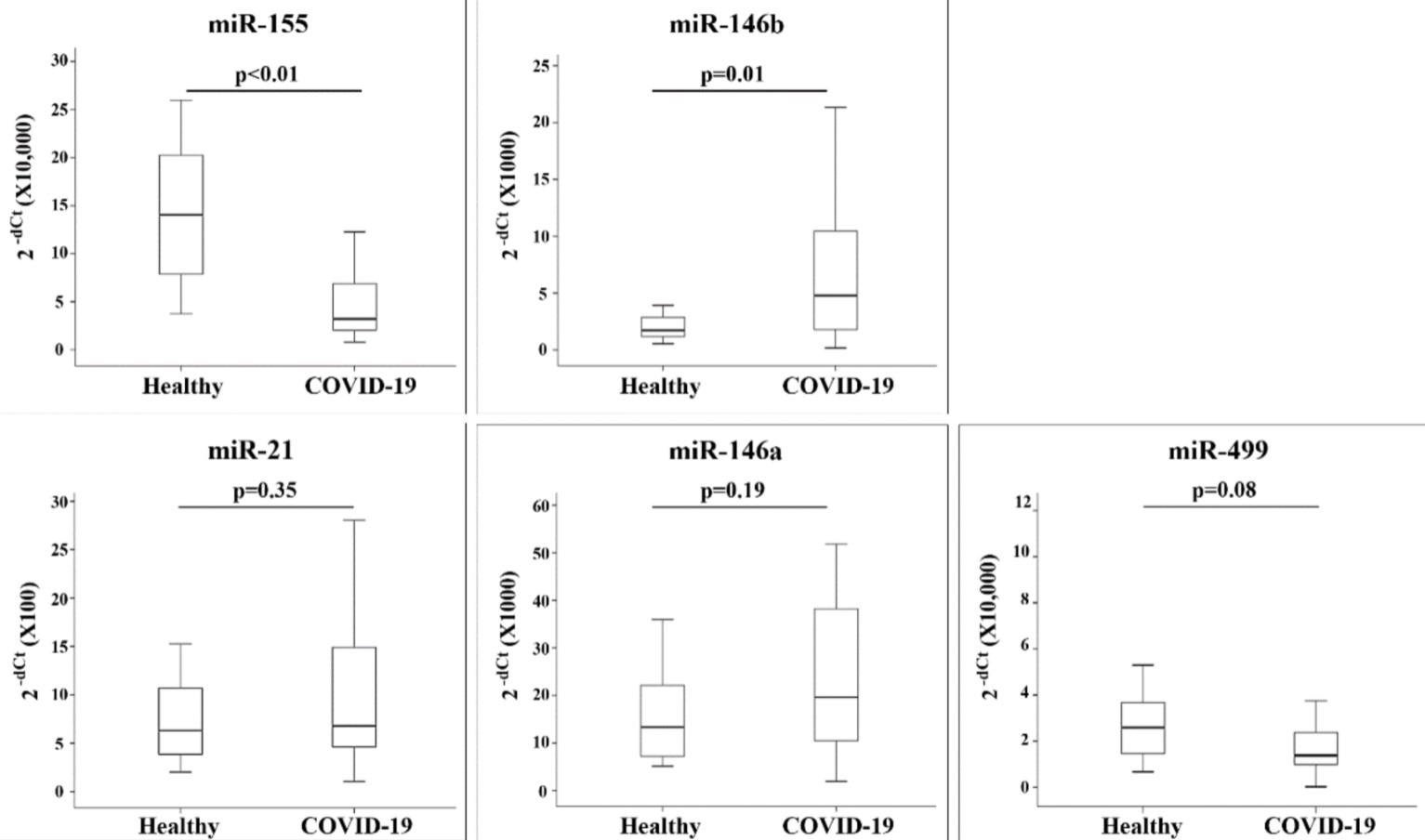 MiRNA expression in healthy and COVID-19 patients: 2−dCt values quantified by quantitative RT-PCR (qRT-PCR) are presented as box plots showing the 10th, 25th, 50th, 75th, and 90th percentile of the patients.
MiRNA expression in healthy and COVID-19 patients: 2−dCt values quantified by quantitative RT-PCR (qRT-PCR) are presented as box plots showing the 10th, 25th, 50th, 75th, and 90th percentile of the patients.
Prediction of clinical course
The levels of circulating miR-155 and miR-146b were estimated in patients who died during hospitalization and in those who survived. This was done to determine whether these miRNAs can be used as prognostic biomarkers. All patients who died were mechanically ventilated and were hospitalized for a longer duration than those who survived.
A significantly lower expression of miR-155 was observed in patients who died compared to that in patients who survived. Further statistical analysis revealed that a polymerase chain reaction (PCR0 cycle threshold value (Ct value) of 1.98 can be used as a cut-off to predict the chances of death and survival among hospitalized COVID-19 patients. The Ct value obtained from PCR reaction inversely correlates with the expression of miRNAs.
Specifically, the analysis revealed that while a Ct value of more than 1.98 predicts survival, a value lower than 1.98 predicts mortality. Notably, the value showed a sensitivity of 84% and a specificity of 50%.
Study significance
The study identifies miR-155 as a prognostic biomarker that shows high sensitivity and specificity in predicting the chances of survival and death in hospitalized COVID-19 patients. As mentioned by the scientists, estimation of circulating miR-155 during hospitalization can be beneficial for identifying at-risk patients and facilitating therapeutic decision-making.
- Kassif-Lerner R. 2022. miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients. Journal of Personalized Medicine. https://www.mdpi.com/2075-4426/12/2/324
Posted in: Medical Research News | Medical Condition News | Disease/Infection News
Tags: Biomarker, Blood, Coronavirus, Coronavirus Disease COVID-19, CT, Immune System, Medicine, Molecule, Mortality, Nucleotides, Oxygen, Pathogen, Personalized Medicine, Polymerase, Polymerase Chain Reaction, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome

Written by
Dr. Sanchari Sinha Dutta
Dr. Sanchari Sinha Dutta is a science communicator who believes in spreading the power of science in every corner of the world. She has a Bachelor of Science (B.Sc.) degree and a Master's of Science (M.Sc.) in biology and human physiology. Following her Master's degree, Sanchari went on to study a Ph.D. in human physiology. She has authored more than 10 original research articles, all of which have been published in world renowned international journals.
Source: Read Full Article


