Scientists have for the first time identified the structure of a protein fiber linked to early-onset type 2 diabetes.
Amylin is a protein that regulates the body’s glucose levels. This small peptide hormone can aggregate or clump together to form amyloid fibrils (a fiber-like structure). These aggregates are a hallmark of type 2 diabetes—although scientists do not yet know precisely how amyloid formation causes the disease.
The formation of amyloid fibrils is also linked to other diseases including the neurodegenerative disorders Alzheimer’s, Parkinson’s and Huntingdon’s Disease.
In research published today in the journal Nature Structural and Molecular Biology, scientists at the University of Leeds report that they have been able to visualize the structure of amylin fibrils using the latest electron microscope technology—and have discovered an architecture that they suspect makes some amylin sequences more prone to form amylin aggregates than others: a feature linked to earlier onset of type 2 diabetes.
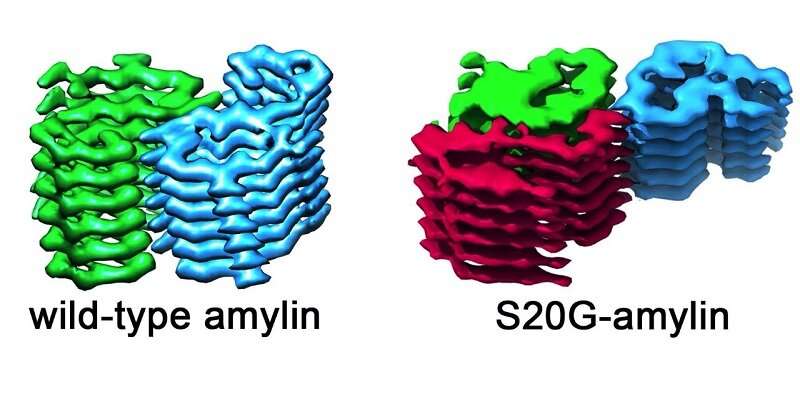
They compared amylin fibrils taken from the type of amylin found in most of the human population—scientists call this the wild-type—and compared it with a genetic variant called S20G observed in people who have early-onset type 2 diabetes.
By analyzing thousands of images, they were able to visualize how amylin molecules aggregate into fibrils, forming an intricate structure in which amylin molecules stack up like rungs in a ladder, and to describe the detailed interactions that hold the fibrils together.
The images revealed that the fibrils formed by the wild-type and S20G versions of amylin are different. All of the wild-type fibrils have two copies of amylin per rung. This was also true for some of the S20G fibrils but crucially, they also found a form of the S20G fibrils with three amylin fibrils per layer. This suggests that fibrils can form templates onto which more copies of amylin can lock.
This could account for why the S20G-variant protein aggregates more quickly and is associated with more rapid onset of disease.
Neil Ranson, Professor of Structural Molecular Biology and Deputy Director of the Astbury Centre for Structural Molecular Biology at the University of Leeds, co-led the project. He said: “This is a really exciting result because it reveals a mechanism for how ever-larger aggregates might form and that is crucial in understanding the disease process.
“We know this happens in disease but we have never understood clearly how it happens. Now with these structures we’re getting the first glimpse what might be going on.”
Sheena Radford FRS, Astbury Professor of Biophysics and Director of the Astbury Centre and co-lead for the study, added: “We know that the S20G protein aggregates more quickly, and this study provides the rationale as to why that might be the case. This is important, not just for understanding amylin—but for understanding many amyloid diseases in which run-away fibril formation occurs.”
The researchers used the technique of cryo-electron microscopy to reveal the structure of the fibrils. The protein sample was frozen and then analyzed in an electron microscope to a resolution where you can see individual atoms.
The electron microscopes are housed within the Astbury Biostructure Laboratory, a major international research centre for structural biology established in 2016 with funding from the University of Leeds and the Wellcome Trust.
Knowledge of amyloid structures such as those presented in this work may pave the way to better diagnosis and treatment of amyloid diseases by customized treatments dependent on the fibril type formed.
Source: Read Full Article


